VASCo
I recently came across VASCo, a program for the calculation of protein surface properties and the visualization of annotated surfaces. Of course you can do great visualization with it, but not only. It can appear very usefull when you want to study protein-protein interactions inside a given structure, and this is achieved through the surface point distances option. And of course you can also map electrostatic potential or hydrophobicity onto these surface points. And don’t worry about how to display the result in PyMOL, it’s actually super easy thanks to the plugin that comes with the program.
What VASCo can do
VASCo can display surfaces information:
- electrostatic surfaces
- lipophilic (hydrophobic) surfaces
- contact distances between molecules (both in the asymmetric unit and with symmetry mates)
How to install VASCo on Windows
-
Install version 2.7.11 of Python.
-
Go to VASCo’s website. Download the program and request a licence (free for academics). For me it took about 24h before they sent the licence, very quick! Unzip the VASCo archive and execute VASCo-Modules-x.win32.exe, then select the python distribution (2.7.11).
-
Run PyMOL and select Plugin > Install Plugin and select the ppixplugin_vx.py, then restart Pymol.
How to use VASCo
Then for each PDB file you need to generate a .ppix.gz file which contains the info that is used by the plugin to generate the surfaces (electrostatic, lipophilic, contact patch).
How to do that:
-
Put the pdb file and VASCo.py in the same folder. Also create a subfolder called “input” and copy the pdb file inside.
-
Open a terminal (in Windows: Start Menu > cmd) and go to that folder.
-
Type the command:
python VASCo.py –in_dir ./ -filename PDBname.pdbIt will tell you that no option file was found and offer to create one, answer “Yes”. It will create a file called input.ppix which you can further modify if you want (but I leave it untouched, as the default parameters are all good for me). -
Type the same command again, this time it will read the input.ppix and run VASCo further.
Then VASCo runs for a few minutes. The bigger your PDB file, the longer the calculation will take. A PDB file with 800 amino acids takes about 3 minutes on my computer. The biggest file I have run so far was 4000 amino acids and it took 90 minutes.
Visualizing the output
The important file will be located in subdirectory output/PDBname/ppixdb_out/ and will be called PDBname_db.ppix.gz.
Then start Pymol, open your pdb file, then go to Plugin > Vasco surface loader > open file > PDBname_db.ppix.gz
Tadaaah!
For each molecular properties you want to visualize you will need to reload by ticking the appropiate boxes. Read the Documentation for more info.
VASCo vs APBS
What I like about VASCo compared to APBS is that you can change the colours as you wish. Also the files are much smaller that APBS: a few Mo for VASCo, a few hundreds of Mo for APBS.
A few images
If you are not convinced yet, here are a few images using PDB 5BO6 as an example:
Electrosatic surface display:
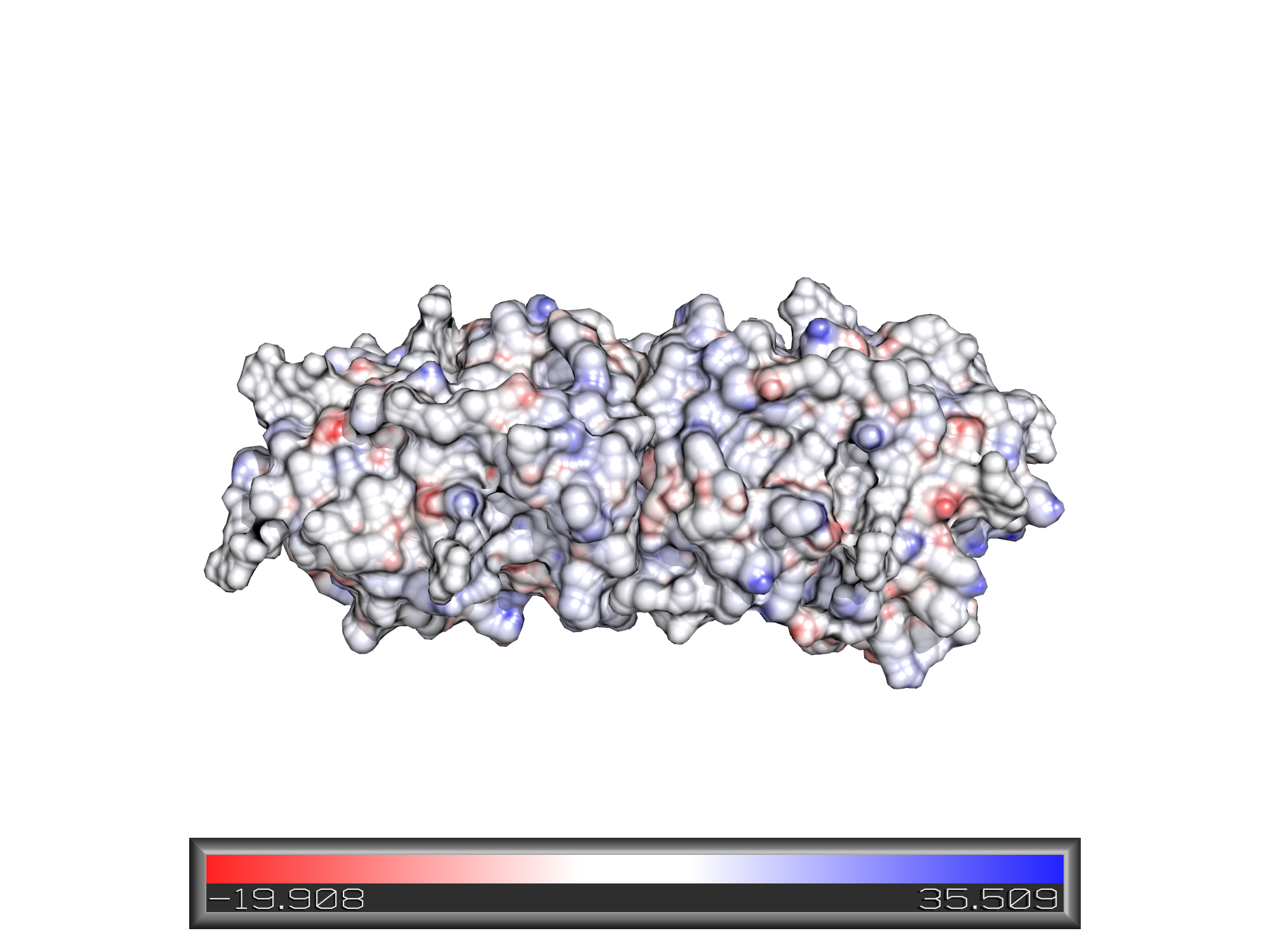
Lipophilic surface display:
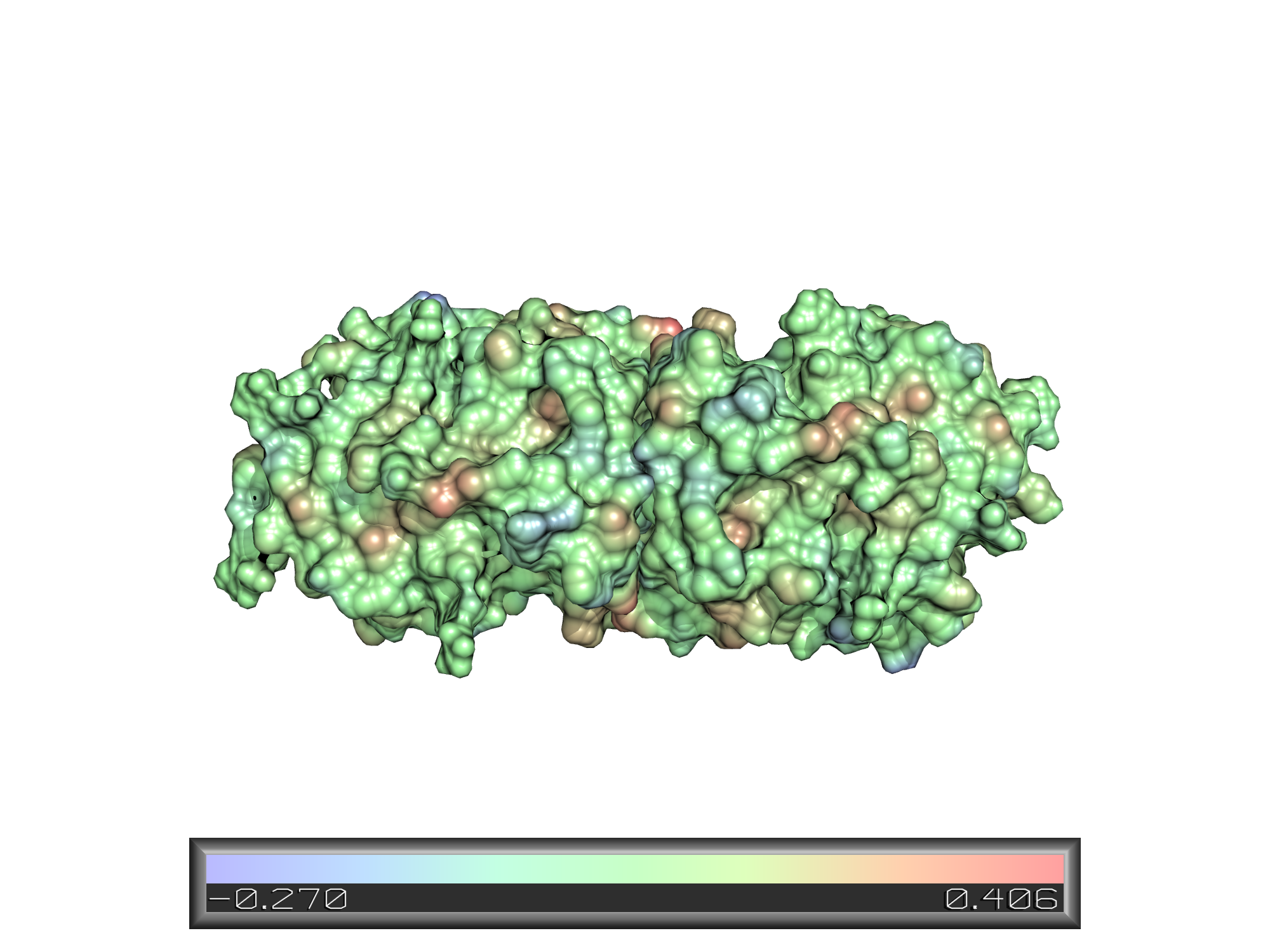
Patch distance display:
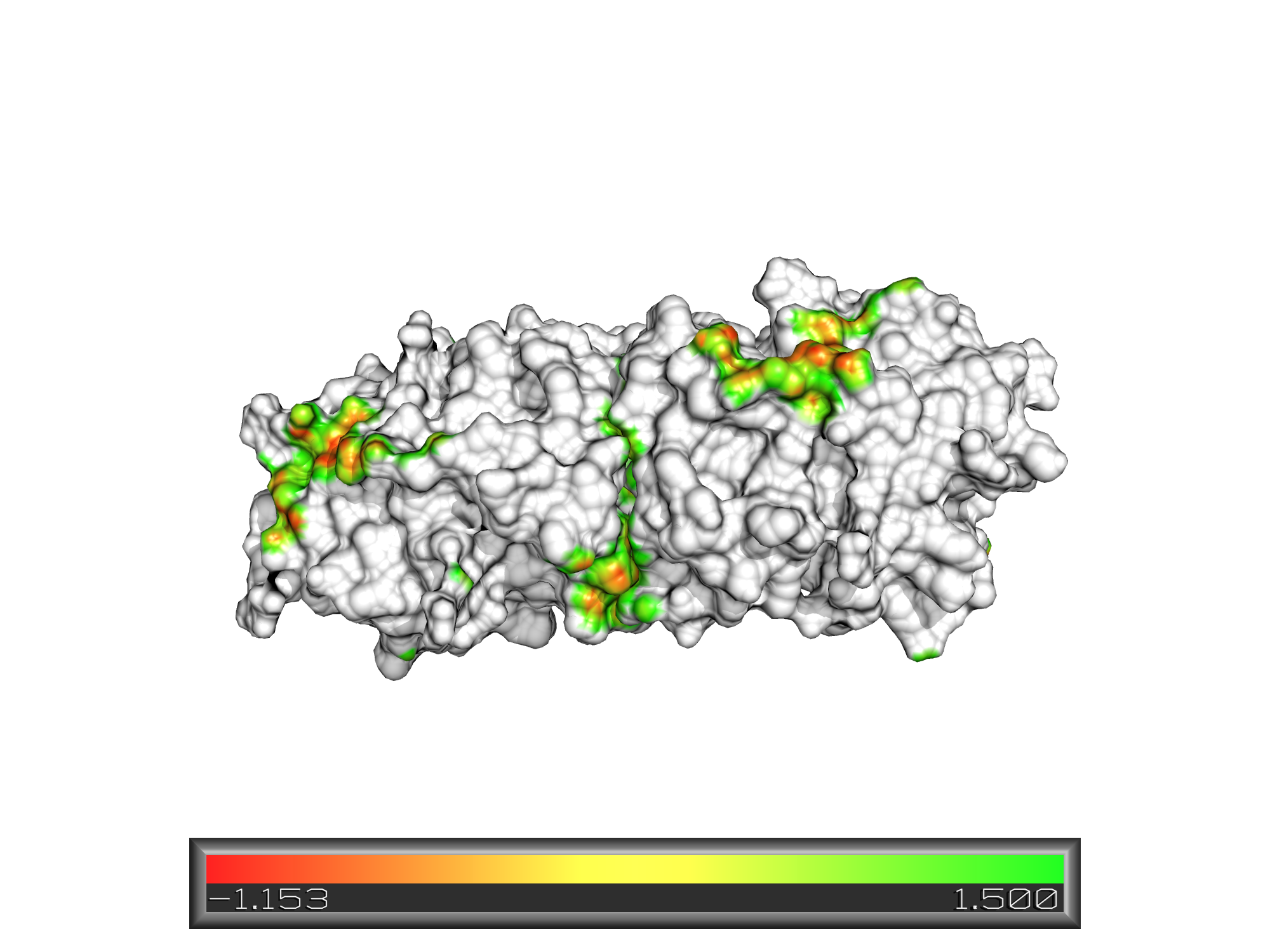
Patch distance display with symmetry mates:
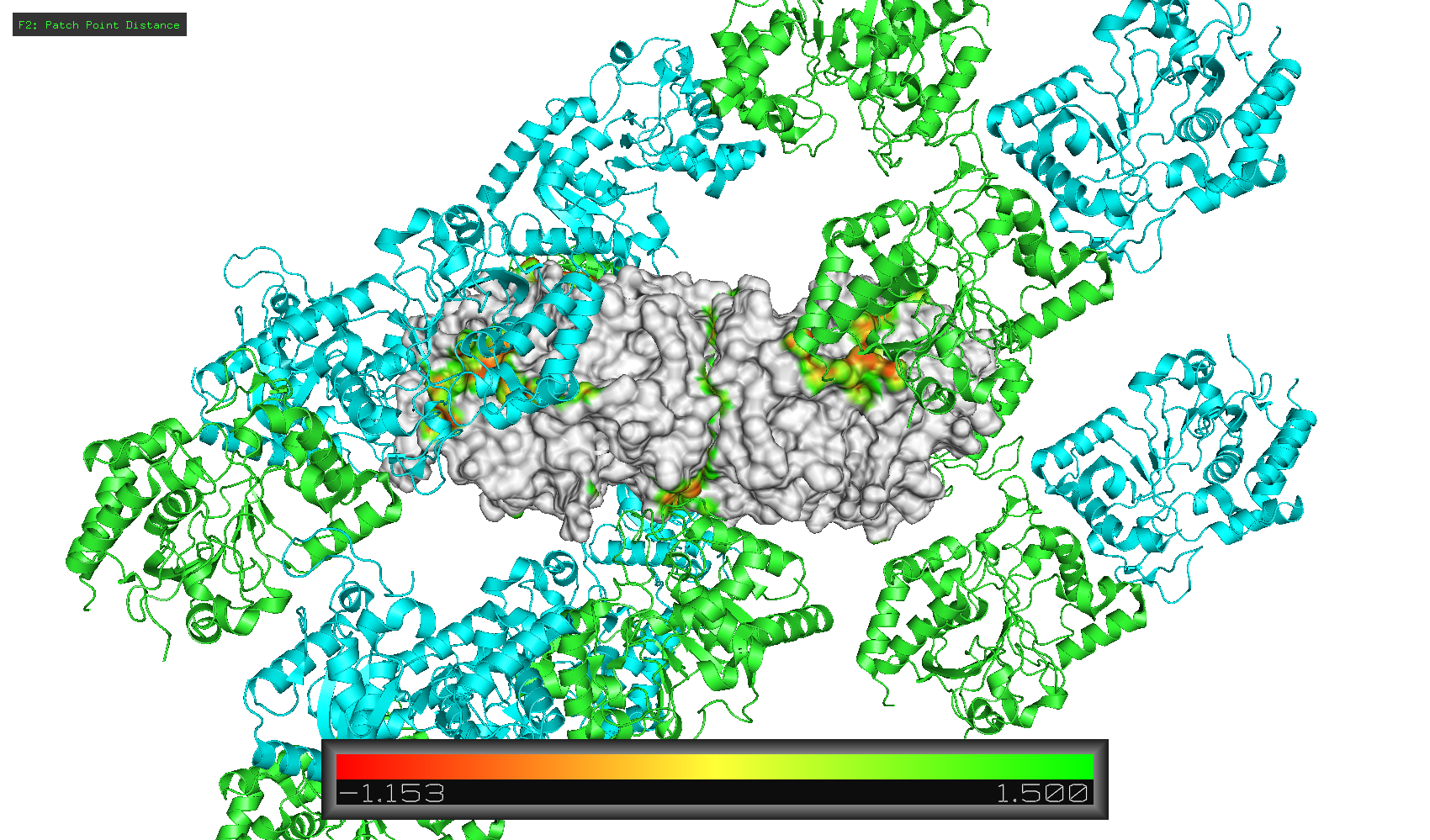
Cheers!
DH ~(‾▿‾)~
 Tui Lab
Tui Lab